Cenik Lab
Overview
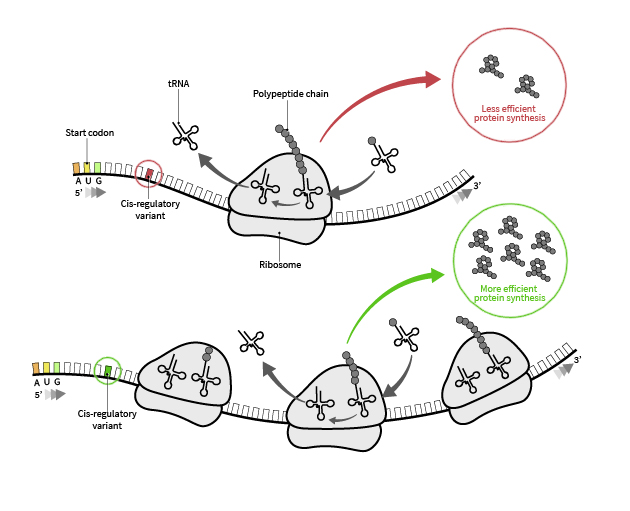
Our long-term goal is to develop the necessary computational and experimental framework for predictive models that explain how cells determine their protein abundance. Achieving this goal involves two major components: (1) higher-resolution and higher-precision measurements of various gene expression modalities, and (2) computational and theoretical advancements capable of integrating these quantitative measurements into cohesive, predictive frameworks.
Our Research Interests
Single cell ribosome profiling for characterizing translation in development, cancer and immunology
We developed a novel microfluidic isotachophoresis method (Ribo-ITP) to measure ribosome occupancy from ultra-low input samples including single cells.
Learn MoreA computational framework to understand translational control
We develop machine learning models to integrate quantitative measurements of translation into cohesive, predictive frameworks.
Learn More