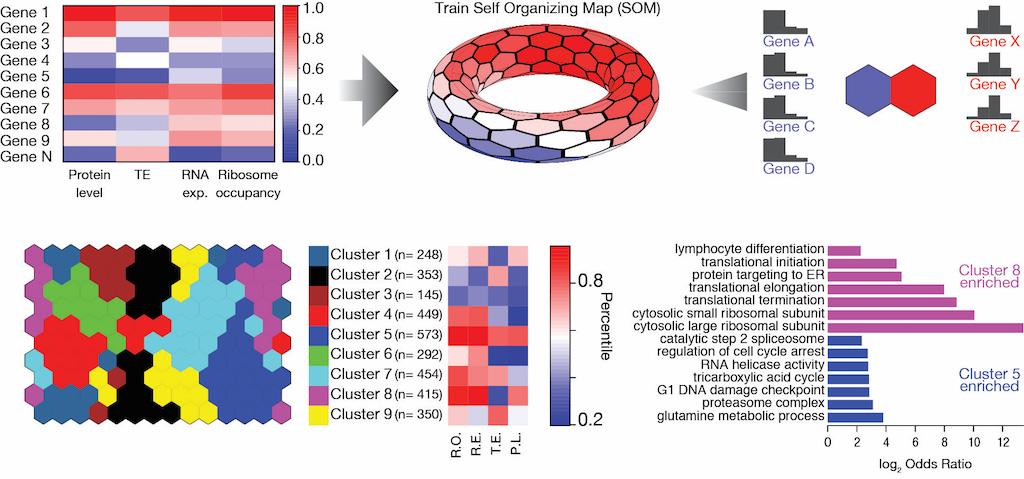
Translation efficiency covariation across cell types is a conserved organizing principle of mammalian transcriptomes
Liu Y, Rao S, Hoskins I, Geng M, Zhao Q, Chacko J, Ghatpande V, Qi K, Persyn L, Wang J, Zheng D, Zhong Y, Park D, Cenik ES, Agarwal V, Ozadam H, Cenik C§
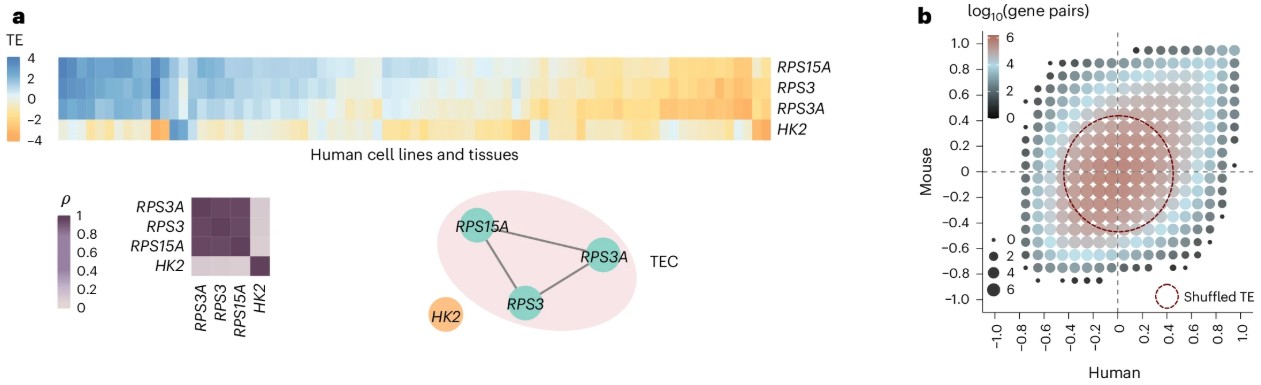
Predicting the translation efficiency of messenger RNA in mammalian cells.
Zheng D*, Persyn L*, Wang J*, Liu Y, Montoya FU, Cenik C§, Agarwal V§

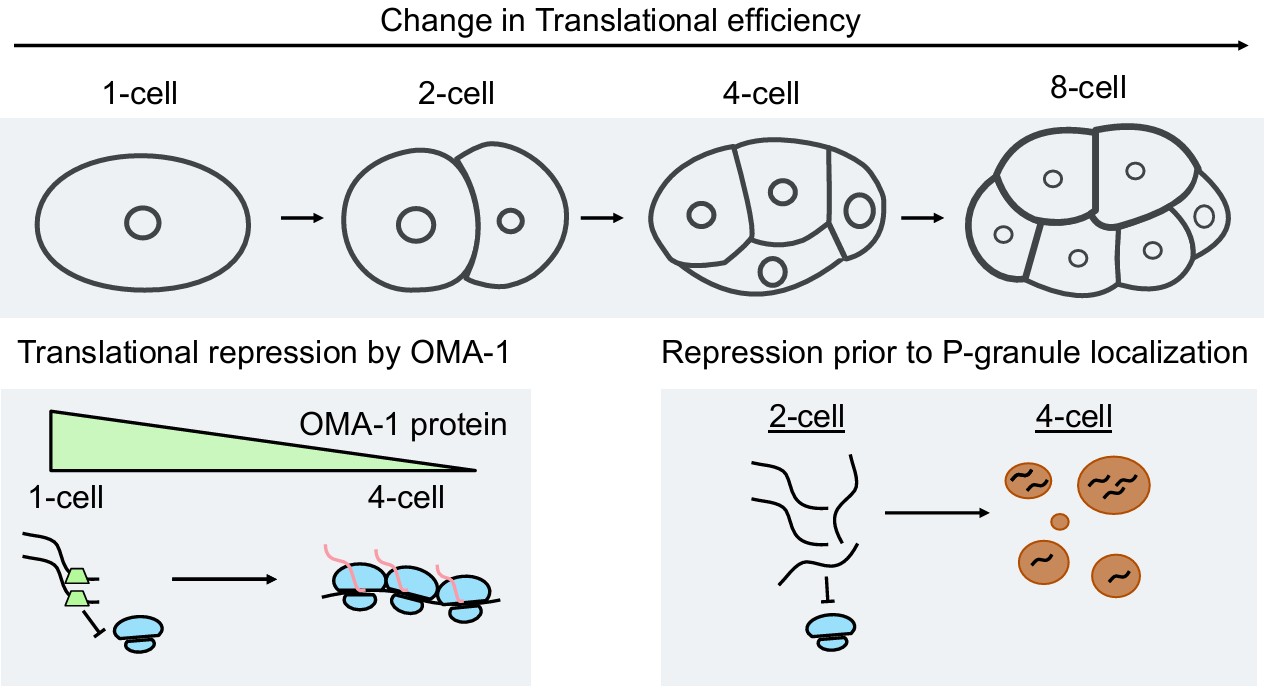
Landscape and regulation of mRNA translation in the early C. elegans embryo
Shukla Y, Ghatpande V, Hu CF, Dickinson DJ§, Cenik C§
A programmed decline in ribosome levels governs human early neurodevelopment
Ni C, Wei Y, Vona B, Park D, Wei Y, Schmitz DA, Ding Y, Sakurai M, Ballard E, Li L, Liu Y, Kumar A, Xing C, Qin S, Kim S, Foglizzo M, Zhao J, Kim HG, Ekmekci C, Karimiani EG, Imannezhad S, Eghbal F, Badv RS, Schwaibold EMC, Dehghani M, Mehrjardi MYV, Metanat Z, Eslamiyeh H, Khouj E, Alhajj SMN, Chedrawi A, Ramzan K, Hashmi JA, Alluqmani MM, Basit S, Veltra D, Marinakis NM, Niotakis G, Vorgia P, Sofocleous C, Lee H, Jeong WC, Umair M, Bilal M, Alves CAPF, Sieber M, Kruer M, Houlden H, Alkuraya FS, Zeqiraj E, Greenberg RA, Cenik C, Yu L, Maroofian R, Wu J, Buszczak M
Cycloheximide resistant ribosomes reveal adaptive translation dynamics in C. elegans
Zhao Q, Bolton B, Rothe R, Tachibana R, Cenik C, Sarinay Cenik E
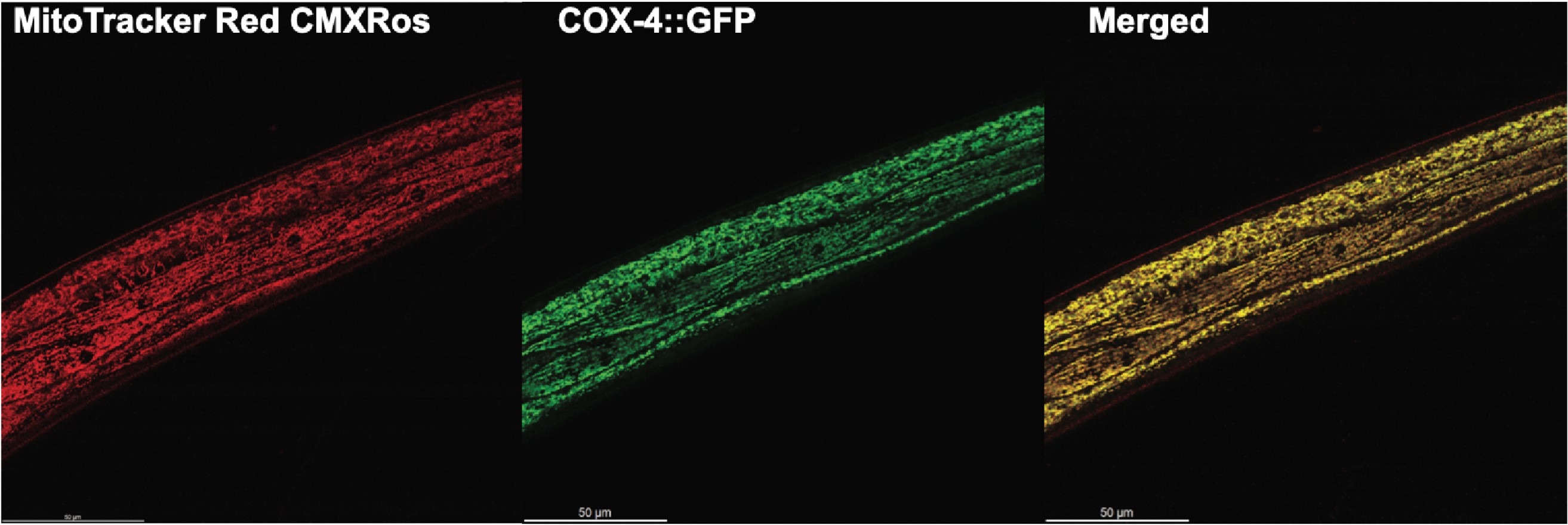
Differential impacts of ribosomal protein haploinsufficiency on mitochondrial function
Surya A, Bolton B, Rothe R, Mejia-Trujillo R, Leonita A, Zhao Q, Liu Y, Rangan R, Gorusu Y, Nguyen P, Cenik C, Sarinay Cenik E
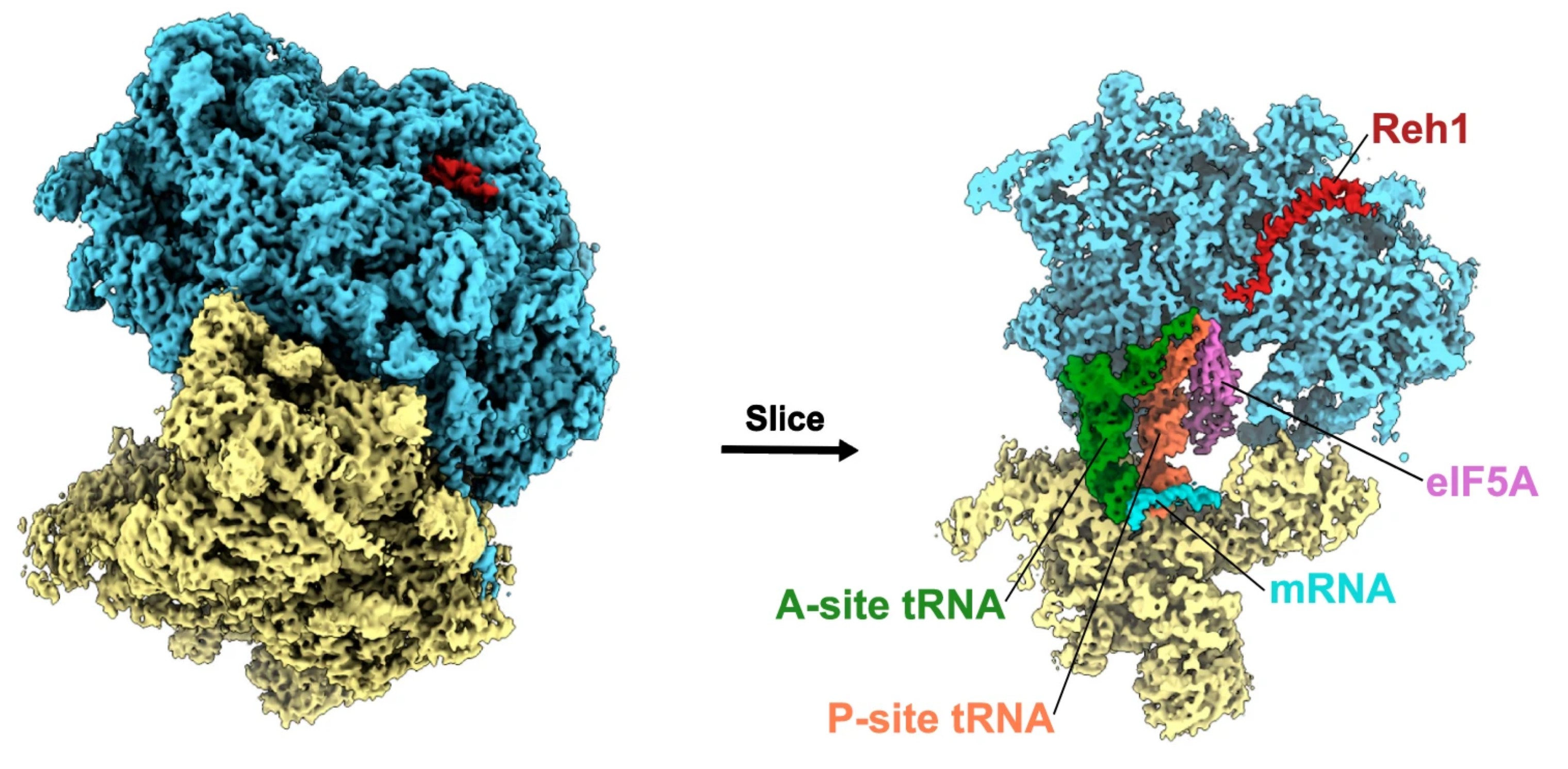
The assembly factor Reh1 is released from the ribosome during its initial round of translation
Musalgaonkar S, Yelland J, Chitale R, Rao S, Ozadam H, Taylor D, Cenik C§, Johnson AW§.
Long-read RNA sequencing reveals allele-specific N 6-methyladenosine modifications
Dayea Park, Cenik C§

RiboGraph an interactive visualization system for ribosome profiling data at read length resolution
Chacko J*, Ozadam H*, Cenik C§
Integrated multiplexed assays of variant effect reveal determinants of catechol-O-methyltransferase gene expression
Ian Hoskins, Shilpa Rao, Charisma Tante, Cenik C§
Single cell quantification of ribosome occupancy in early mouse development
Ozadam H*, Tonn T*, Han C*, Segura A, Hoskins I, Rao S, Ghatpande V, Tran D, Catoe D, Salit M, Cenik C§
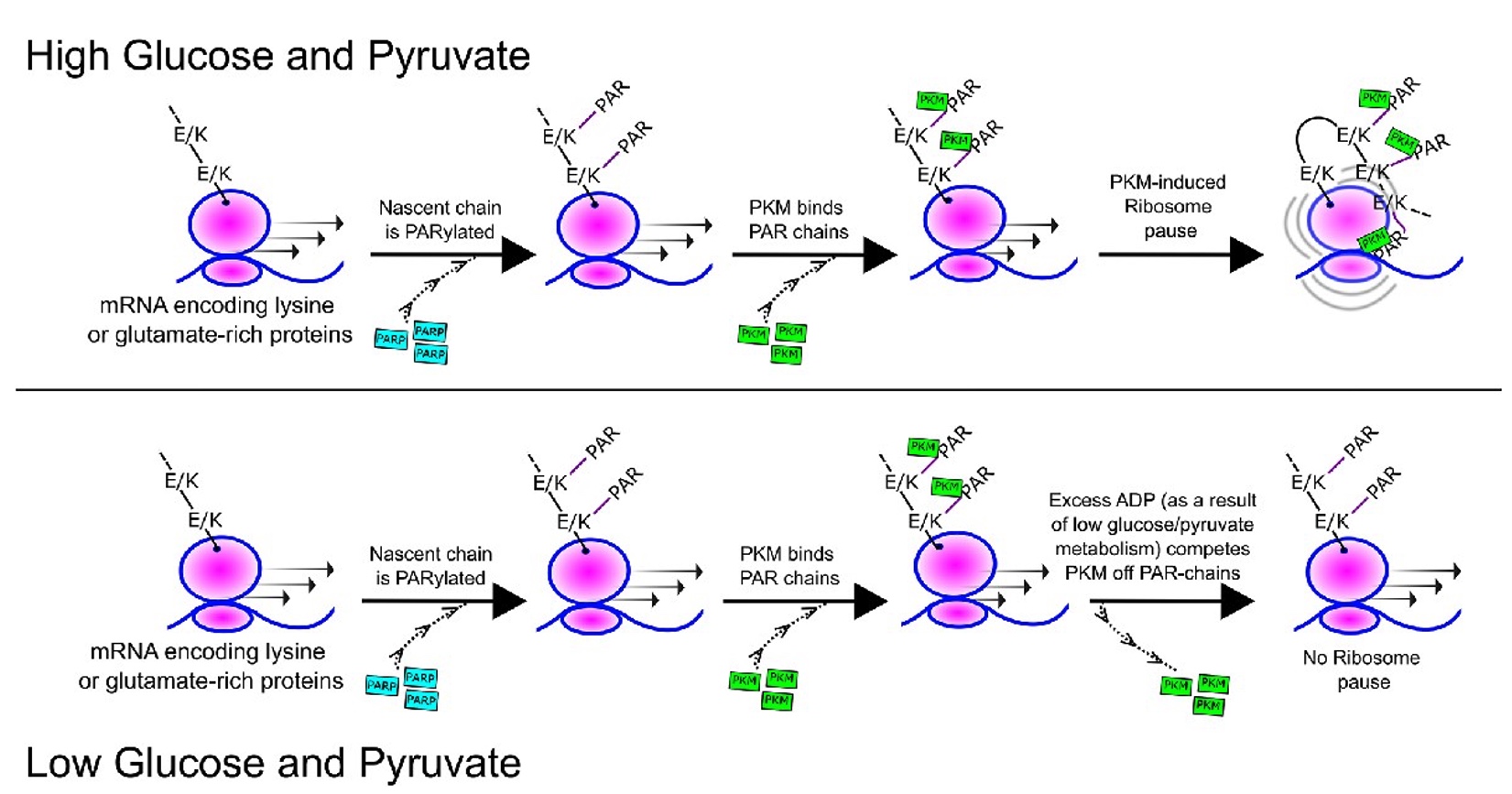
Pyruvate Kinase M (PKM) binds ribosomes in a poly-ADP ribosylation dependent manner to induce translational stalling
Kejiou NS, Ilan L, Aigner S, Luo E, Tonn T, Ozadam H, Lee M, Cole GB, Rabano I, Rajakulendran N, Yee BA, Najafabadi HS, Moraes TF, Angers S, Yeo GW, Cenik C, Palazzo AF
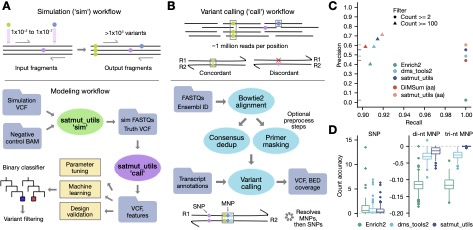
satmut_utils: a simulation and variant calling package for multiplexed assays of variant effect
Hoskins I, Sun S, Cote A, Roth FP Cenik C§
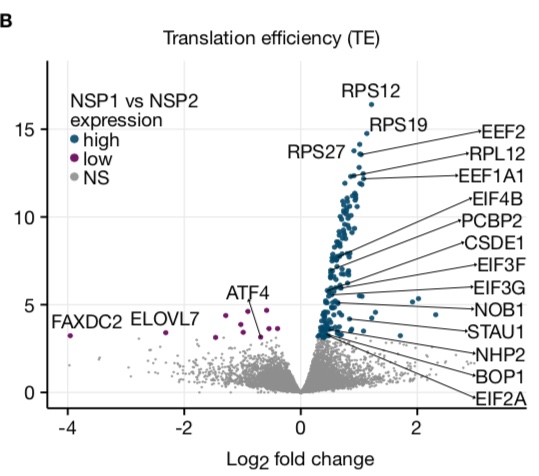
Genes with 5' terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 NSP1 protein
Rao S, Hoskins I, Garcia PD, Tonn T, Ozadam H, Cenik ES, Cenik C§
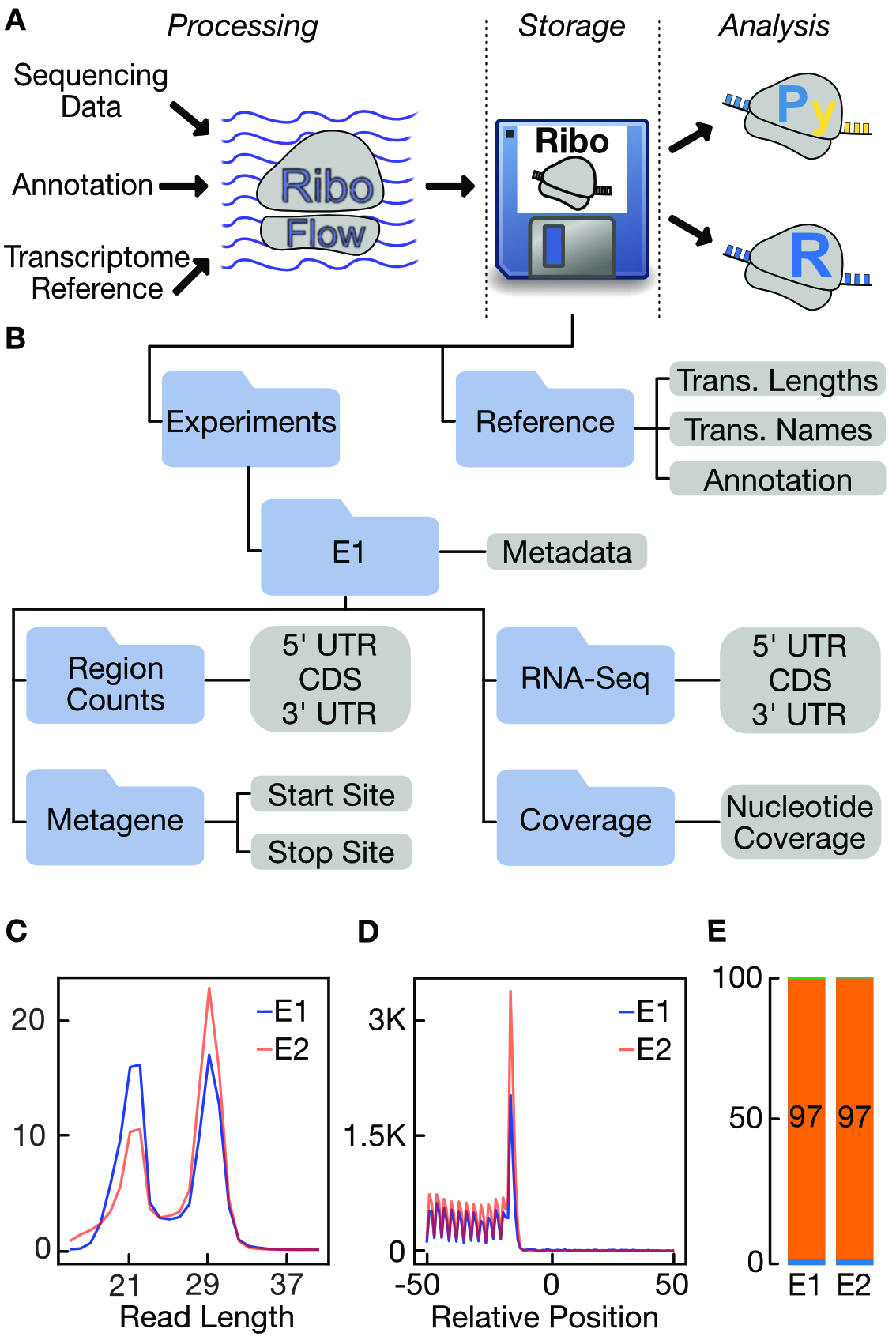
RiboFlow, RiboR and RiboPy: An ecosystem for analyzing ribosome profiling data at read length resolution
Ozadam H, Geng M, Cenik C§
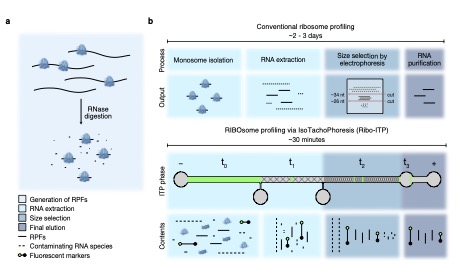
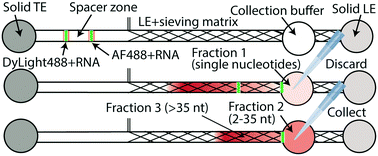
Simultaneous RNA purification and size selection using on-chip isotachophoresis with an ionic spacer
Han CM, Catoe D, Munro SA, Khnouf R, Snyder MP, Santiago JG, Salit ML, Cenik C§
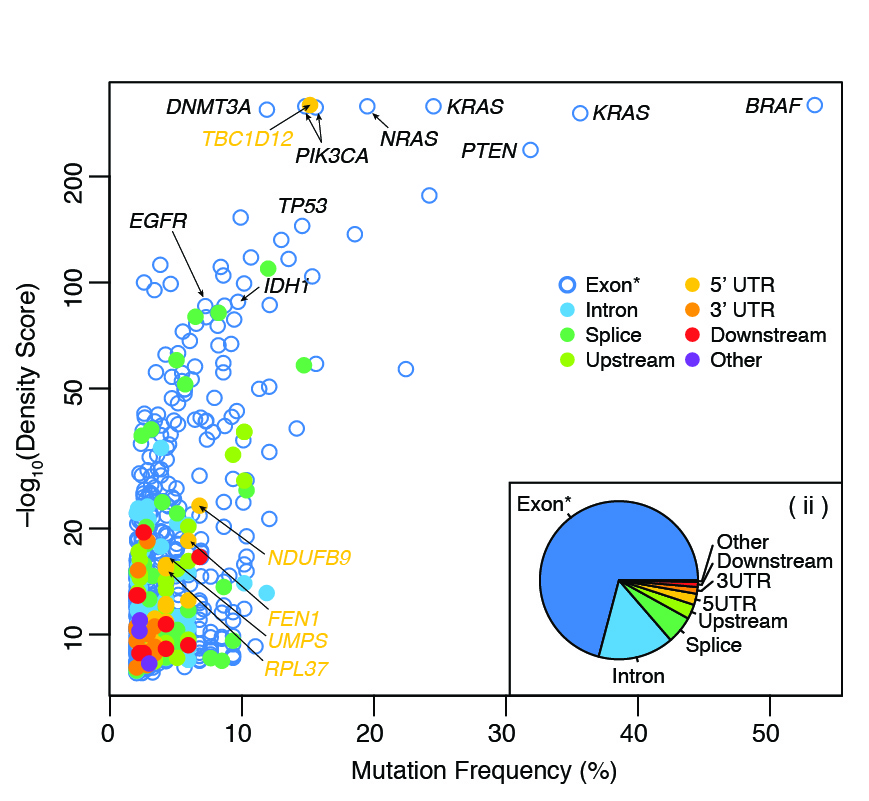
Systematic identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations
Araya CL*, Cenik C*, Reuter JA, Kiss G, Pande VS, Snyder MP, Greenleaf WG
Integrative analysis of RNA, translation and protein levels reveals distinct regulatory variation across humans
Cenik C, Cenik ES, Byeon GW, Grubert F, Candille S, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, Ricci E, Snyder MP